Lipid MS-MS spectra data
Lipidomics Bulkhttps://www.thermofisher.com/fr/fr/home/industrial/mass-spectrometry/mass-spectrometry-learning-center/mass-spectrometry-applications-area/metabolomics-mass-spectrometry/lipidomics-workflows.html
Lipid MS/MS spectra are acquired through tandem mass spectrometry, where lipids are first ionized and then fragmented to produce characteristic ions. Mass-to-charge ratios (m/z) and intensities of these fragment ions are detected and recorded as MS/MS spectra. The spectra are then analyzed using software to match observed fragments with theoretical lipid structures, enabling lipid identification and quantification.
Production systems

nanoHPLC-Exploris 480 coupling
This device offers very high acquisition speed, high resolution (4800000) and allows the use of different acquisition modes(Full-Scan, ddMS2, DIA, PRM). It is used to analyse very complex protein mixtures (identification and quantification).
See more about the technology
µHPLC-Q-Exactive plus coupling
This instrument offers high-speed, high-resolution acquisition (240000) and a range of acquisition modes (Full-Scan, ddMS2, PRM, pseudo DIA). It is used to analyse highly complex lipid/metabolite mixtures (identification and quantification).
See more about the technologyAssociated pipelines
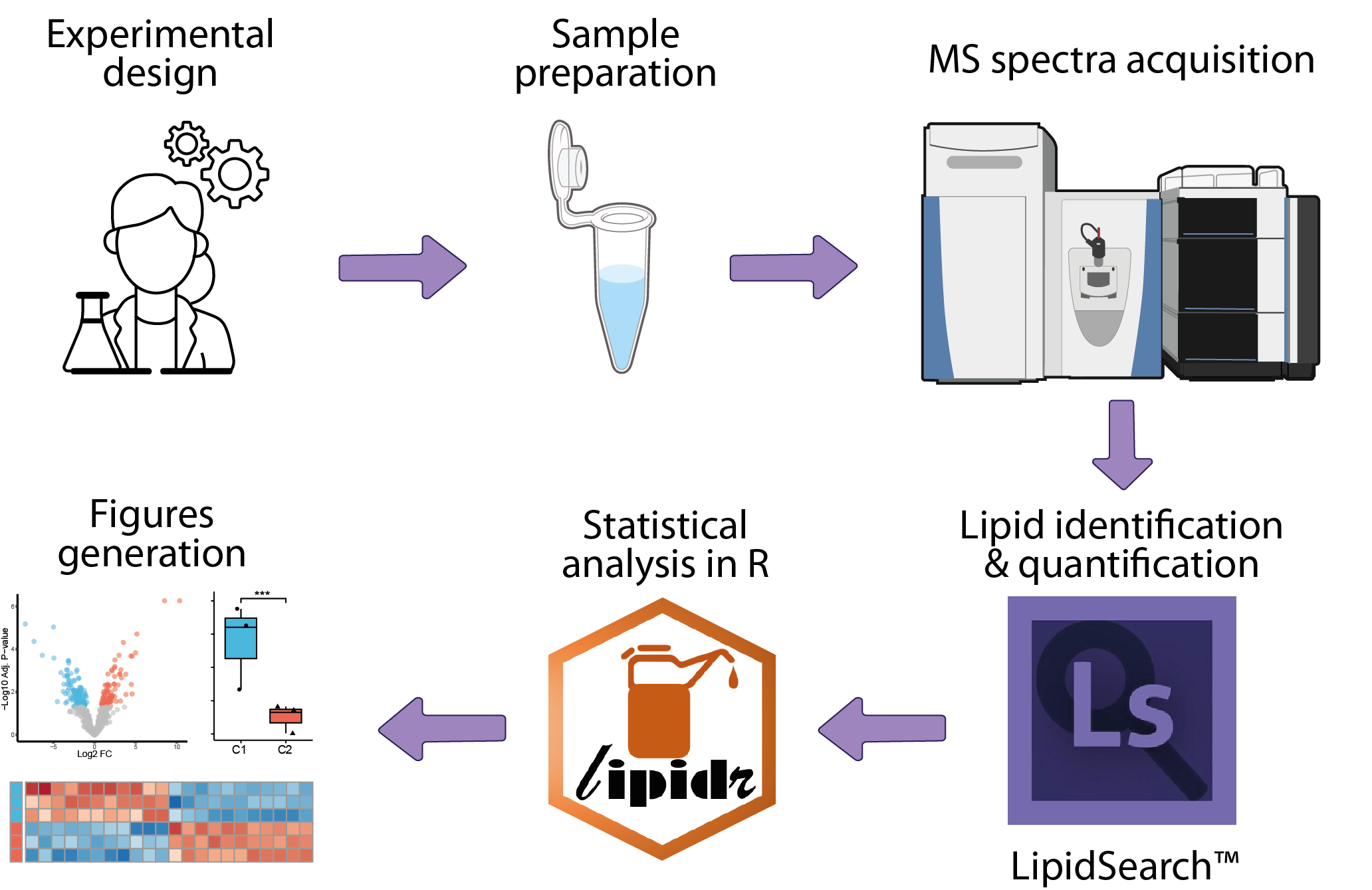
mass-spectrometry-based lipidomics
The analysis of mass-spectrometry-based lipidomics data begins with the collection of raw mass spectrometry data. The next step involves lipid identification, where spectra are matched to known lipid molecules using specialized algorithms, followed by lipid quantification. This primary analysis is typically conducted by the platform managing the mass spectrometer, utilizing dedicated software such as LipidSearch. The resulting data can then be statistically analyzed within an R framework to identify lipids differentially quantified between conditions.
Go to pipeline documentation

