Documented pipelines
The main goal of ODIN portal is to store documented pipelines that are used to analyse omics data. The documentation covers the type and resolution of quantitative omics data to which the pipeline is dedicated, which production systems are able to generate them, and which platform manages this system, before describing the actual bioinformatics analysis steps. Each step in the data processing required to extract biological information is contained in a dedicated module, characterized by its inputs and outputs, as well as the computing environment (IDE, softwares, packages) in which it can be run on the network's IT infrastructures.. The code associated with each module is presented in the form of a notebook, where it is supplemented by guidelines and graphical representations to help users understand the procedure. Finally, publications in which the pipeline has been used are referenced as examples of what can be achieved.
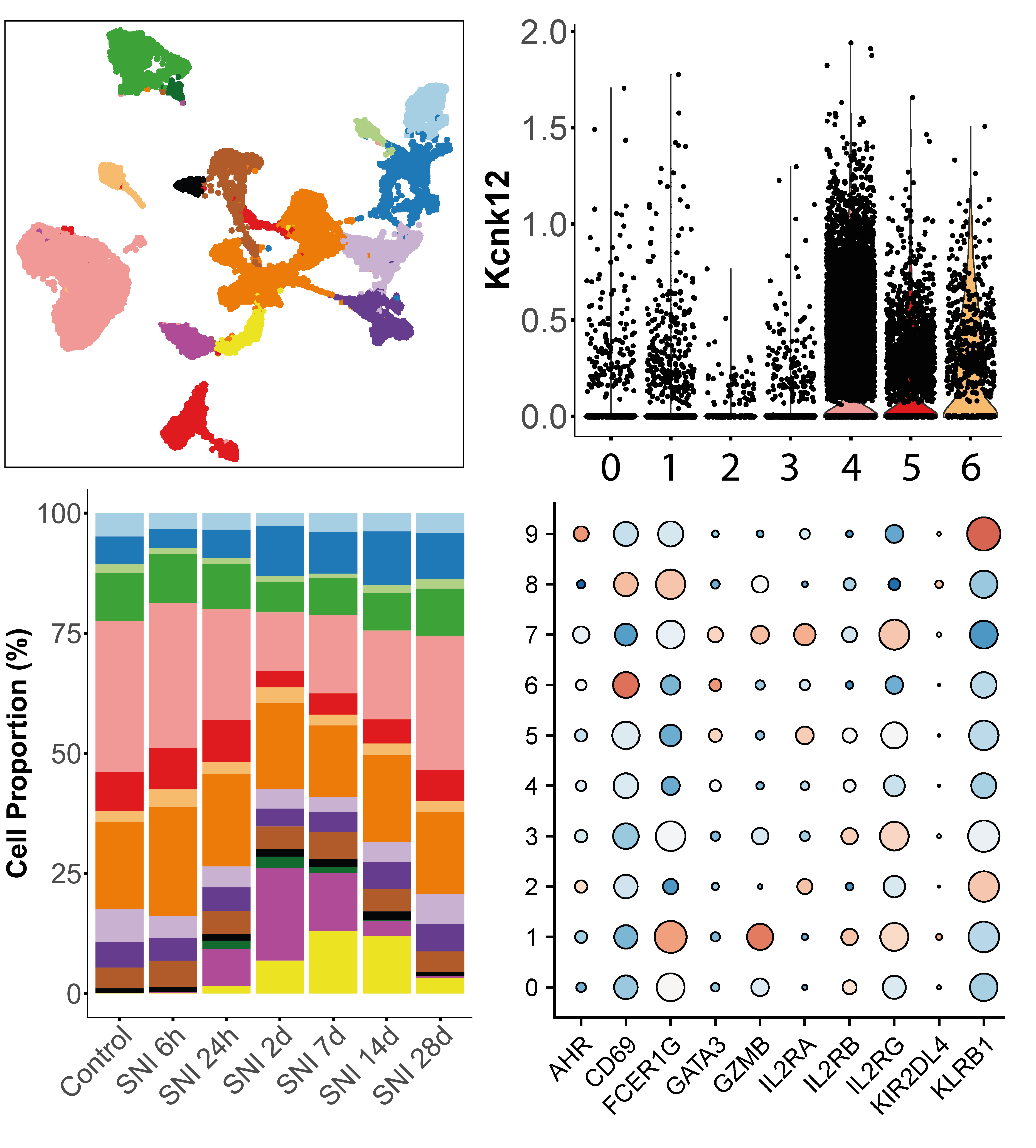
droplet-based single-cell RNA-seq
Analysis of single-cell gene expression data generated by droplet-based systems
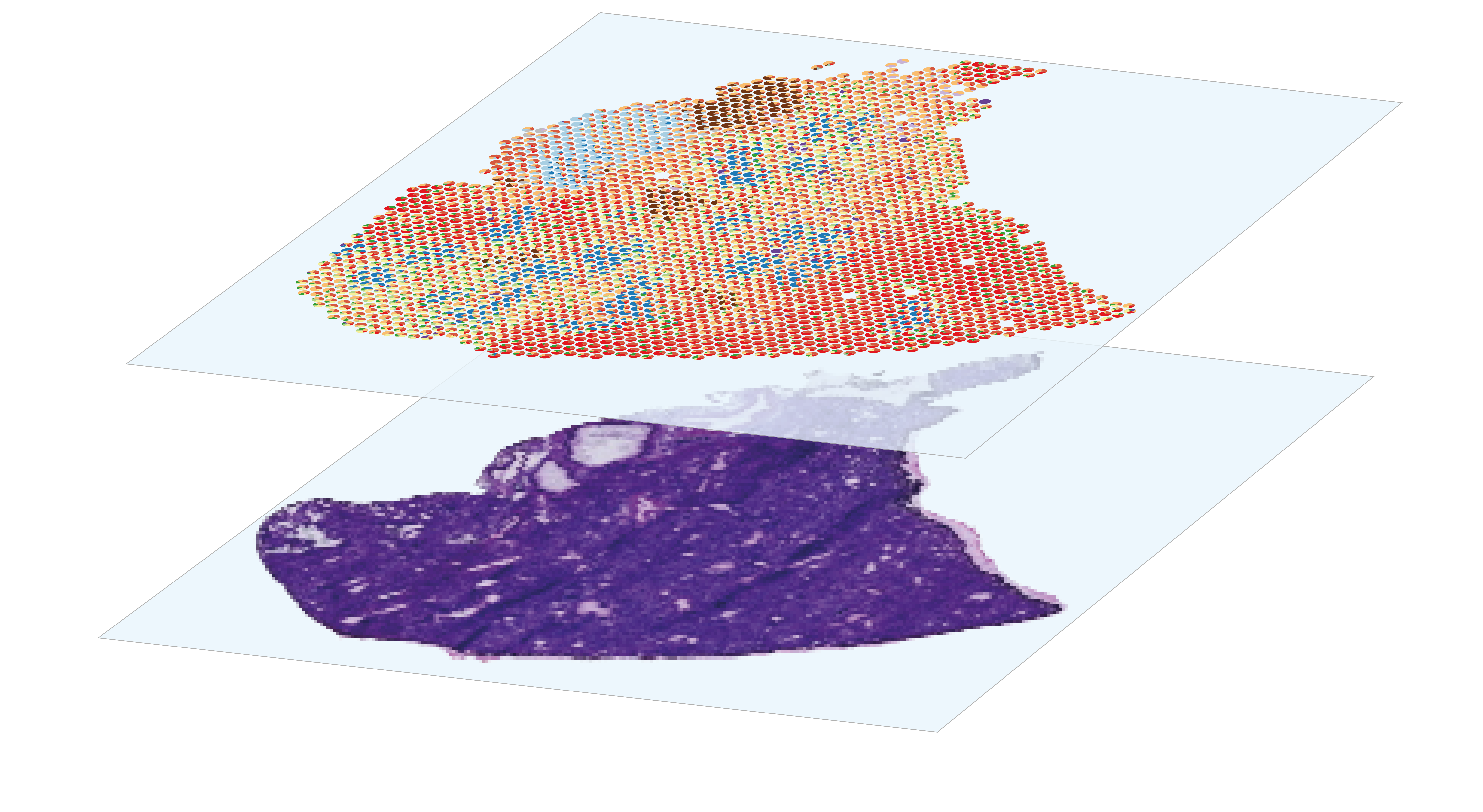
sequencing-based spatial transcriptomics
Analysis of spatial gene expression data generated by sequencing-based systems

imaging-based in-situ spatial single-cell
Analysis of single-cell gene expression data generated by imaging-based in-situ systems
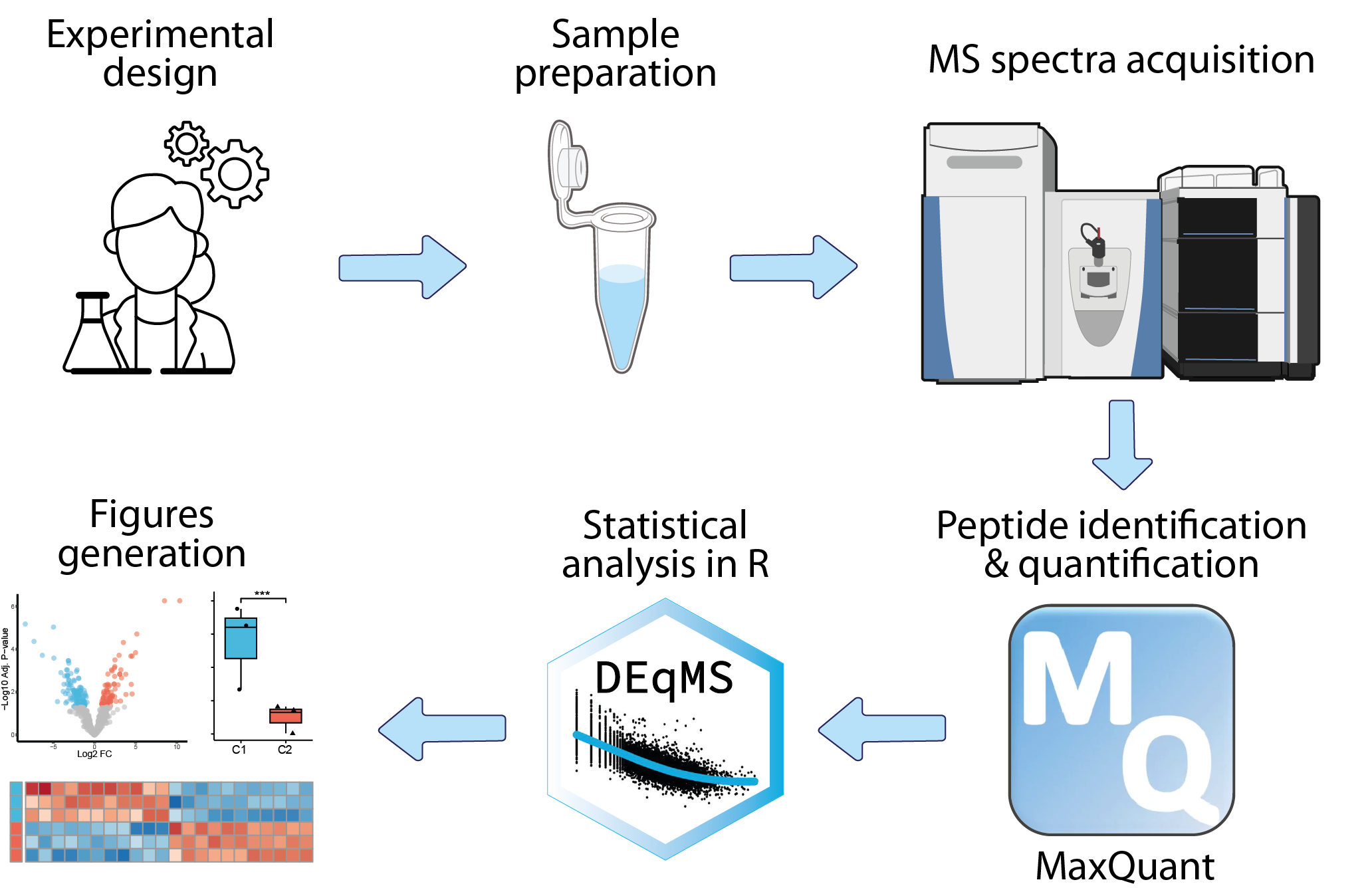
mass-spectrometry-based proteomics
Analysis of mass-spectrometry-based proteomics data
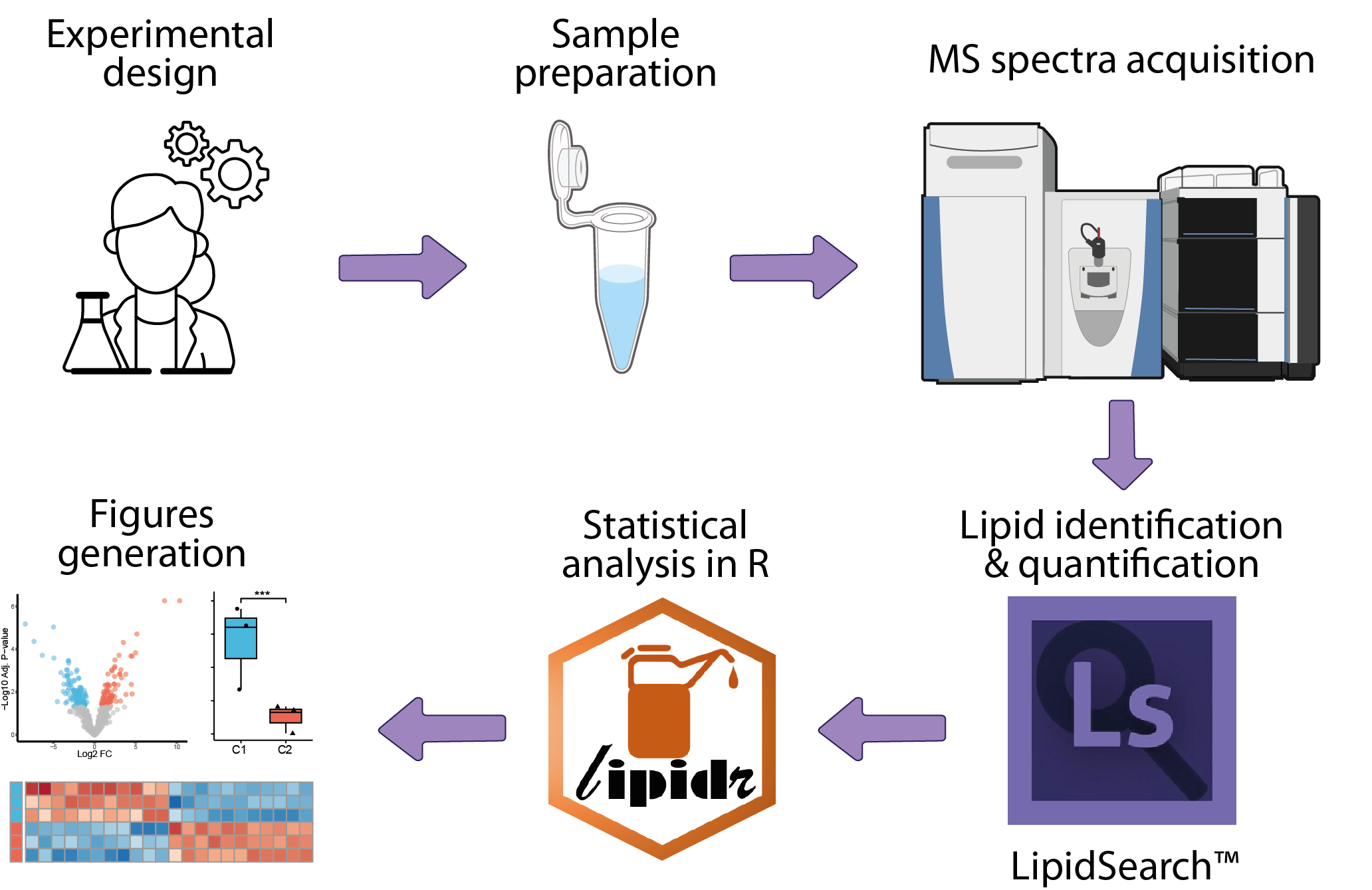
mass-spectrometry-based lipidomics
Analysis of mass-spectrometry-based lipidomics data
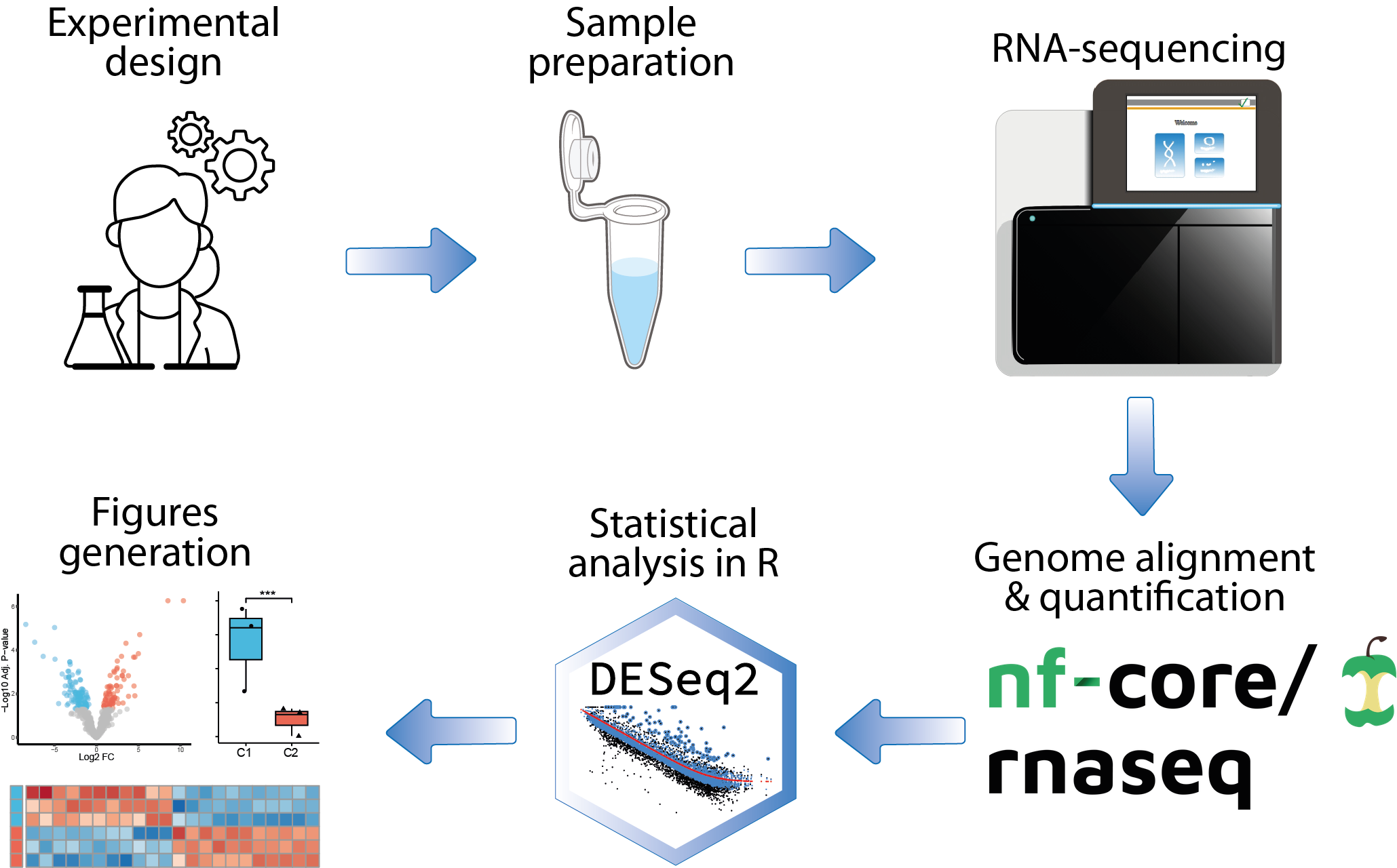
bulk RNA-seq
Analysis of bulk gene expression data generated by RNA-seq
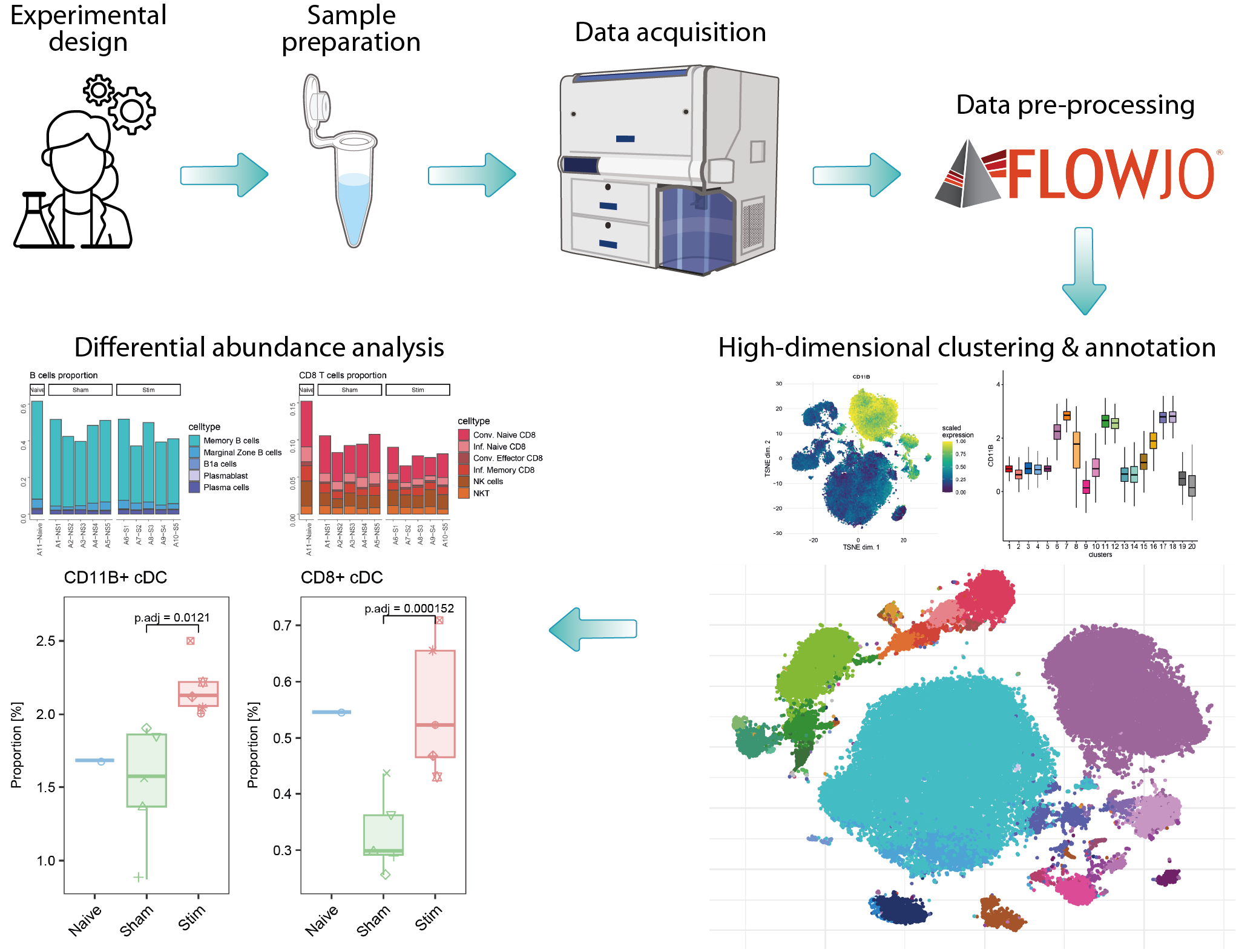
high-dimensional cytometry
Analysis of high-dimensional cytometry data generated by flow or mass cytometry systems


