Here we describe an automatic labeling procedure using scanvi and/or scMusketeers that relies on mixing spatial cells and a home-made or external single-cell reference cells into a unique latent space to predict spatial cells cell type. Obviously the single-cell reference needs to be as close as possible as the spatial dataset in term of cell type occurence.
scanvi automatic labeling
Home-made or external single-cell reference used for transfert labeling needs to be prepared before automatic labeling could be performed. Reference AnnData object needs to have a layer 'counts' filled with either raw or log-normalized counts and a cell type field (ClusterName for zeisel et al., 2018) in the .obs metadata. On bego, some references are already available for mouse brain dataset based on 2 references publications:
* Yao et al. (2023)
* Zeisel et al. (2018)
import scanpy as sc
import scispy as scis
# Yao et al. (2023) 10xGenomics mouse brain 247 genes panel
scref_yao2023 = sc.read_h5ad('~/000-Atlas-references/mousebrain_247g_raw_4M_10p.h5ad')
scref_yao2023.layers['counts'] = scref_yao2023.X.copy()
# Zeisel et al. (2018) - whole CNS
scref_zeisel2018 = sc.read_h5ad('~/000-Atlas-references/zeisel_2018_CNS.h5ad')
# Zeisel et al. (2018) - coronal section
scref_zeisel2018 = sc.read_h5ad('~/000-Atlas-references/zeisel_2018_coronal.h5ad')
scref_zeisel2018.layers['counts'] = scref_zeisel2018.raw.X.copy()
# read your spatial dataset
ad6_harmony = sc.read_h5ad('000-outs/ad6.h5ad')
# transfert annotation
scis.pp.scvi_annotate(ad6_harmony, scref_zeisel2018, batch_size=127, label_ref='ClusterName', label_key='scanvi', metaref2add=['TaxonomyRank1','TaxonomyRank2','TaxonomyRank3','TaxonomyRank4','Region'])
ad6_harmony.var_names = ad6_harmony.raw.var_names
# save your annotate dataset
ad6_harmony.write('000-outs/ad6_scanvi.h5ad')
scMusketeers automatic labeling
import scanpy as sc
# parameters
class_key = "scmusk"
unlabeled_category = "Unknown"
batch_key = "sample"
h5ad_ref = "~/000-Atlas-references/zeisel_2018_coronal.h5ad"
h5ad_query = '000-outs/ad6_scanvi.h5ad'
# prepare ref
ref = sc.read_h5ad(h5ad_ref)
ref.obs = ref.obs.reset_index(drop=True)
ref.obs.index = ref.obs.index.map(str)
ref.X = ref.raw.X.copy()
ref.obs[class_key] = ref.obs['ClusterName']
# arrange a random batch
s = [random.randrange(1, 4) for _ in range(0, ref.shape[0])]
ref.obs[batch_key] = s
ref.obs[batch_key] = ref.obs[batch_key].map(str)
ref.var_names_make_unique()
ref.obs_names_make_unique()
ref.write('ref.h5ad')
# prepare query
query = sc.read_h5ad(h5ad_query)
query.obs = query.obs.reset_index(drop=True)
query.obs.index = query.obs.index.map(str)
query.X = query.layers['counts'].copy()
query.obs[class_key] = unlabeled_category
query.var_names_make_unique()
query.obs_names_make_unique()
query.write('query.h5ad')
scMusketeers need to be run as command line within a specific conda environnement on bego
conda activate /data/analysis/ML_models/conda_env/cb_scmusketeers
sc-musketeers transfer ref.h5ad --query_path query.h5ad --class_key=scmusk --unlabeled_category=Unknown --batch_key=sample --out_dir=. --out_name=scmusk.pred
Then get back to python notebook
import anndata as ad
pred = sc.read_h5ad("scmusk.pred.h5ad")
pred = pred[pred.obs.train_split == 'test']
pred.obs.index = query.obs.index
query.obs['scmusk'] = adata_pred.obs['scmusk_pred']
query.obs['scmusk'] = query.obs['scmusk'].astype("category")
query.write('000-outs/ad6_scanvi_scmusk.h5ad')
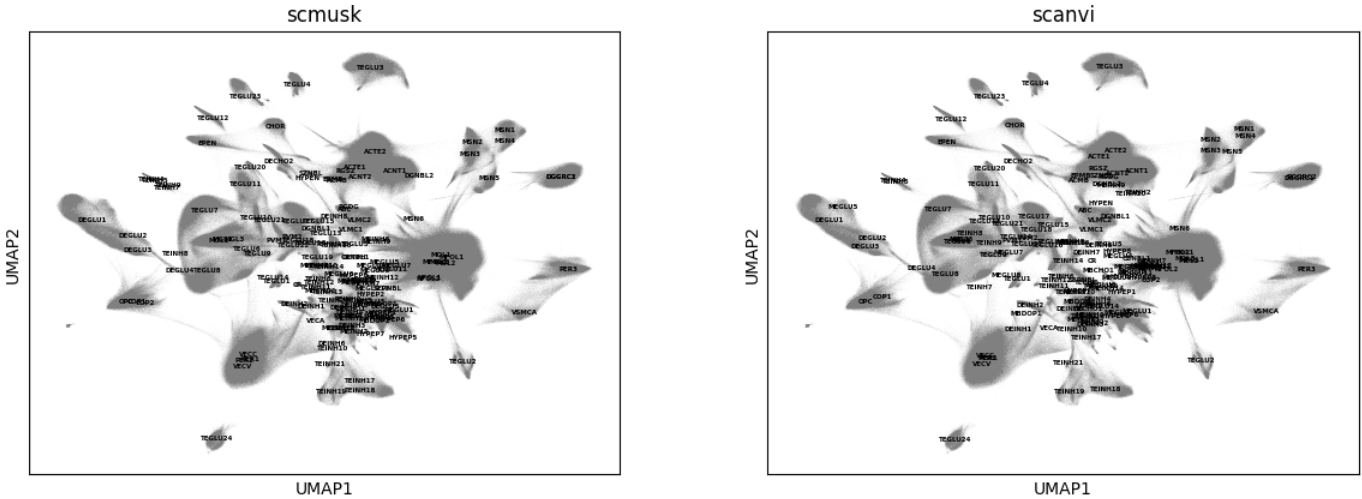
Visualize UMAP by cell type
sc.pl.embedding(query, "umap", color=['scmusk','scanvi'], legend_loc='on data', legend_fontsize=4)
Re-export metadata for xenium explorer
samples = pd.read_csv('samples.csv', sep=',')
for index, row in samples.iterrows():
sdata = SpatialData.read("000-outs/SD_" + row['sample'])
sub = query[query.obs['sample'] == row['sample']]
sub.uns["spatialdata_attrs"] = sdata.table.uns["spatialdata_attrs"]
sub.obs.cell_id = sub.obs.cell_id.astype(str)
del sdata.tables["table"]
sdata.tables["table"] = sub
# xenium case
spatialdata_xenium_explorer.write("000-outs/XE_" + row['sample'], sdata, shapes_key='cell_boundaries', points_key='transcripts', image_key='morphology_focus', polygon_max_vertices=40, mode='+o')